Drug-susceptibility tests in tuberculosis: contradictory or neglected?
DOI:
https://doi.org/10.5935/2764-734X.e20250459Keywords:
Tuberculosis, Multidrug-Resistant, Real-Time Polymerase Chain Reaction, Genetic Testing, Case ReportsAbstract
Tuberculosis continues to have a negative impact on public health, especially its multidrug-resistant variant. Consequently, rapid molecular tests are used to identify the genetic mutations responsible for resistance to the first- and second-line drugs used for treatment. However, these tests have detection limitations in some clinical cases. Therefore, all available information must be collected and validated. Here, we report a case where the laboratory results showed discrepancies between molecular and phenotypic methods—an aspect that had not been considered in the case’s conclusion. This led to the discussion regarding noncompliance with established clinical care algorithms, which deserve to be revisited.
Downloads
INTRODUCTION
Although Brazil is not considered a high-burden country for multidrug-resistant tuberculosis, the underdetection of the disease and the unsatisfactory outcomes of its treatment in recent years are a real threat to the disease’s elimination in our country1. Therefore, one of the goals to which Brazil committed at the first high-level meeting on tuberculosis held in 2018 by the World Health Organization (WHO) was to make new and better tests available for point-of-care diagnosis and to carry out drug sensitivity tests for the rapid diagnosis of cases with tuberculosis resistant to the primary drugs used in treatment1.
Consequently, molecular techniques have emerged, especially those based on polymerase chain reaction (PCR), to identify the DNA of the Mycobacterium tuberculosis complex. In addition to being a rapid molecular test (RMT) for the diagnosis of tuberculosis, it identifies mutations in the rpoB gene that confer resistance to rifampicin. Another test available is the Line Probe Assay (LPA), which detects mutations that confer resistance not only to rifampicin, but also to other drugs used in first- and second-line treatment2, 3, 4. The commercial tests most widely used in our country are the Xpert MTB/RIF® Ultra® (Cepheid) and GenoType MTBDRplus® (Hain Lifescience) tests.
However, there are rare mutations outside the target region of these two tests, which may remain undetected5, 6, 7. Considering this limitation, the Brazilian Ministry of Health recommends that the two tests be used for the rapid screening for rifampicin resistance, and mycobacteria cultures must be used as a complementary method, especially in patients living with HIV/AIDS (PLWHA) or those who are not progressing well in treatment4, 6.
We report a case where conflicting results between the different M. tuberculosis sensitivity tests were obtained and interpreted late, consequently revealing failure to comply with established clinical care algorithms, which deserve to be revisited.
CASE REPORT
The patient was a 27 year old transexual woman, a former sex worker who had been incarcerated for 8 years and was released about 30 days before our first appointment. Her medical history dated back 5 months when she had a cough with yellowish sputum, fever in the afternoon (not measured), night sweats, dyspnea on medium exertion, and unquantified weight loss. She was diagnosed with tuberculosis in the penal institution based on RMT, without detection of resistance to rifampicin. The date of this first exam was designated as “D1” in the chronological sequence that will be used henceforth. Following the initial tuberculosis diagnosis, the patient began treatment with RHZE (5 fixed-dose combination tablets of rifampicin 150 mg + isoniazid 75 mg + pyrazinamide 400 mg + ethambutol 275 mg) for 2 months. She proceeded to the second phase of treatment, with daily doses of rifampicin 750 mg and isoniazid 375 mg for another 4 months, completing a total of six months of treatment on D180, while still imprisoned. She progressed with weight gain and improved dyspnea, and ceased to have fever soon after; although, she remained with a productive cough throughout this period. She said he had only had a chest X-ray and a single HIV test (which was negative) at the beginning of the treatment. Moreover, we did not have access to her medical records from the prison system.
Thirty days after the end of the tuberculosis treatment and already released, the patient began to have episodes of afternoon fever (not measured), night sweats, and weight loss, and was referred from the basic health unit to our specialized outpatient department at D210. On reviewing the patient’s tests available on the Laboratory System Manager (GAL) of the Central Public Health Laboratory (LACEN), we found that the same sample collected on D1 had been sent for LPA testing, the result of which was released on D73. The LPA test showed a mutation in the inhA gene and none in the katG and rpoB genes, and the report read “sensitive to rifampicin and resistant to isoniazid.” The same sample from D1 was also used for culture in automated liquid culture medium (BD BACTEC Mycobacterium Growth Indicator Tube - MGIT 960®), which showed M. tuberculosis growth, and a sensitivity test (SIRE®) showed resistance to isoniazid and rifampicin and sensitivity to streptomycin, ethambutol, amikacin, levofloxacin, moxifloxacin, and bedaquiline. These last two results were released on D159.
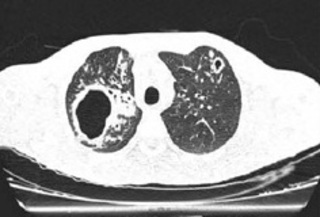
Another important piece of information was obtained from the repetition of the HIV test at a primary care unit, because of the patient’s new complaints, which came back positive. Thus, she was prescribed antiretroviral therapy with tenofovir, lamivudine, and dolutegravir. In a sample taken in the first visit, D210, the HIV viral load count was 29 copies/ml, CD4 T lymphocytes count was 1,360 cells/mm3, and CD8 T lymphocytes count was 513 cells/mm3. On that same day, the patient underwent a chest CT scan, which showed cavitations in both lung apices and areas with a tree-in-bud pattern (Figure 1). In view of the clinical picture, radiological images, and available laboratory test results, the diagnosis of active, multidrug-resistant tuberculosis (MDR-TB) to rifampicin and isoniazid was made. Consequently, the treatment consisted of bedaquiline (400 mg for 14 days followed by 200 mg thrice a week), levofloxacin (1,000 mg once a day), terizidone (750 mg once a day), and clofazimine (100 mg once a day), the latter replacing linezolid, which was in short supply in Brazil in 2024. The smear microscopy of a sputum sample collected on D210 was negative but the RMT was positive, showing no resistance to rifampicin (result released on D211). A second sample collected on D211 showed paucibacillary TB. There was no growth of mycobacteria in the culture of the sputum from D210, nor of the other two samples collected on D240 and D270 (whose smear microscopy results were also negative). Table 1 shows the chronological sequence of all the specific tests and their results, for educational purposes. The patient continues to be monitored on an outpatient basis with good adherence to treatment, no clinical complaints, and radiological improvement.
Figure 1. Tomographic sections performed on D210 showing thick-walled cavitations in both upper lobes (larger on the right), associated with consolidations. There are also some areas with centrilobular opacities, tree-in-bud infiltrates and bronchiectasis, more evident in the right lung.
| Date of collection | Clinical sample | Test performed | Technique or Commercial kit | Results | Release date |
|---|---|---|---|---|---|
| D1 | qPCR | Xpert MTB/RIF Ultra® (Cepheid) | M. tuberculosis complex DNA detected,sensitive to rifampicin | D1 | |
| LPA | GenoType MTBDRplus® (Hain Lifescience) |
inhA gene: mutation detected katG gene: mutation not detected rpoB gene: mutation not detected |
D73 | ||
| Sputum | culture |
BD BACTEC Mycobacterium Growth Indicator Tube (MGIT) 960® |
M. tuberculosis complex | D159 | |
| Sensitivity Test |
BD BACTEC MGIT 960® SIRE® |
Sensitive to streptomycin, ethambutol, amikacin, levofloxacin, moxifloxacin, and bedaquiline. Resistant to rifampicin and isoniazid |
D159 | ||
| AFB test | Ziehl-Neelsen staining | Absence of AFB | D210 | ||
| D210 | Sputum | qPCR | Xpert MTB/RIF Ultra® (Cepheid) | M. tuberculosis complex DNA detected, sensitive to rifampicin | D211 |
| culture | Ogawa-Kudoh solid medium | No growth | - | ||
| D211 | Sputum | AFB test | Ziehl-Neelsen staining | Presence of AFB (1+/4+) | D211 |
| D240 | Sputum | AFB test | Ziehl-Neelsen staining | Absence of AFB | D240 |
| culture | Ogawa-Kudoh solid medium | No growth | - | ||
| AFB test | Ziehl-Neelsen staining | Absence of AFB | D270 | ||
| D270 | Sputum | culture |
BD BACTEC Mycobacterium Growth Indicator Tube (MGIT) 960® |
No growth | - |
DISCUSSION
Sensitivity tests (ST) are essential for detecting resistance to the drugs used to treat tuberculosis. These tests can be performed using phenotypic and genotypic methods, with the latter enabling a faster and more timely diagnosis because they can be performed directly on the initial sputum sample, provided that the sputum smear or RMT are positive3, 4. Conversely, all phenotypic methods (and genotypic methods for extrapulmonary or pulmonary samples other than sputum) can only be carried out using the culture isolate4.
The Xpert MTB/RIF® assay was the first RMT prequalified by the WHO for the diagnosis of resistance in tuberculosis and is based on the amplification of the “Rifampicin Resistance Determining Region” (RRDR) of the rpoB gene of M. tuberculosis. However, a lower sensitivity of the method has been described for paucibacillary samples in general and for detecting the C533G mutation of the aforementioned rpoB gene2. The Ultra® version of Xpert MTB/RIF® was developed to overcome these limitations, as well as to identify the Q513Q and F514F mutations, which are silent mutations3. However, LPA makes it possible to detect or infer mutations in genes other than the rpoB gene, such as inhA and katG for first-line drugs (related to the of resistance primary drugs like rifampicin, isoniazid, and ethionamide) and the gyrA, gyrB, rrs, and eis genes for second-line drugs (related to the resistance to fluoroquinolones and aminoglycosides/cyclic peptides)4.
In our case, although RMT and LPA did not detect resistance to rifampicin, the LPA identified the isoniazid resistance mutation—a result that was available in the first month of the second phase of basic tuberculosis treatment. Moroever, this would have been the usual timing during the treatment to have access to the results of the mycobacteria culture. However, unfortunately, they were released 5 months later. Despite this, the information on isoniazid resistance obtained in the third month of treatment was sufficient to refer the patient to our department, which is a reference center for cases of resistant tuberculosis in the state of Goiás. Additionally, the Brazilian Ministry of Health recommends that “if only an inferred mutation in the inhA gene is identified without a mutation in the katG gene, phenotypic ST for isoniazid and 2nd line ST should be carried out, in addition to starting the drug-resistance regimen according to the resistance profile”4.
We don’t know if any control bacilloscopy was carried out (ideally in the second month of treatment) when the patient was still deprived of her liberty, and she told us that there was no repeat radiological exam. The absence of these two tests can influence cure outcomes. A study conducted in the Brazilian state of Paraíba showed that failure to achieve a cure and abandonment of tuberculosis treatment in the population deprived of liberty are mainly associated with not having a follow-up bacilloscopy and with acquired immunodeficiency syndrome8. These “small” failures, often underestimated, in the management of tuberculosis can have major repercussions on the chain of processes and practices related to patient care, impacting directly or indirectly on the health of the patient and on public health as a whole. In our case, the patient had a satisfactory progress (she gained weight, her dyspnea improved, and she no longer had a fever) despite the inadequate treatment, which is a fairly common situation; however, this does not justify noncompliance with the recommended algorithms6. Finally, the results of the culture and the respective phenotypic ST, although released late, would have been another reason to refer the patient to a reference center before the end of the treatment.
Although we are basically dealing with deficits in the flow of intersectoral communication (e.g., in the sharing of information between the laboratory and the point of care, in this case the prison system), we need to better understand the limitations of laboratory methods. Most of the mutations of interest for identifying tuberculostatic resistance occur in the codons covered by Xpert MTB/RIF® Ultra® and GenoType MTBDRplus®. However, there are other less frequent mutations that can occur outside of the RRDR4. Chan et al. described in 2023 a case of the I572F mutation identified by genetic sequencing of the rpoB gene5, previously identified in Hong Kong, South Africa, and Eswatini (former Swaziland). In the latter country, it is believed that 30% of cases of resistant tuberculosis go undiagnosed because they are attributed to this mutation7. This condition has not been investigated or proven in our case. Genetic sequencing may even be justifiable for epidemiological purposes and for studying resistance profiles in certain countries, but the required time, infrastructure, and investment must be taken into account; ultimately, its cost-benefit is inferior to that achieved by using official algorithms4, 6. Moreover, culture for all tuberculosis cases confirmed by Xpert MTB/RIF® is part of these algorithms, especially in special populations like PLWHA and those deprived of their liberty, for whom sending a sample for culture should be simultaneous with the RMT itself6.
It is to be expected that the management of tuberculosis in the prison system faces obstacles within this system, such as the difficulty of accessing tests in proper laboratories9 and outside it (accessibility to referrals and counter-referrals), or even in relation to the low completeness of compulsory notification data10. This reality alone needs to be addressed more effectively by the authorities to minimize the role of the prison environment as the main determinant of tuberculosis transmission, which has been estimated at 36.9% for prisoners >15 years of age in Brazil11. But it also underlines the need for medical teams to focus on this vulnerable population that is more subject to aggravating factors, such as social, health, economic, behavioral, and cultural inequalities, often overlapped with greater organizational limitations on the health services offered to them1.
CONCLUSION
This case report urges us to understand the importance of recognizing and interpreting the results of drug-susceptibility tests in tuberculosis. Additionally, it highlights the responsibility and commitment of all professionals involved in the diagnostic and care sectors to “always check” this information, which is often delayed by several months. Equally important is the need for professionals to understand and adhere to the protocols and algorithms proposed by the National Tuberculosis Control Program.
References
1. Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde e Ambiente. Boletim Epidemiológico da Tuberculose. Brasília: Ministério da Saúde; 2023 [acesso 2025 mar 29]. Disponível em: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/epidemiologicos/especiais/2023/boletim-epidemiologico-de-tuberculose-numero-especial-mar.2023/view
2. Chakravorty S, Simmons AM, Rowneki M, Parmar H, Cao Y, Ryan J, et al. The New Xpert MTB/RIF Ultra: Improving Detection of Mycobacterium tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing. mBio. 2017 Aug 29;8(4):e00812-7. DOI: 10.1128/mBio.00812-17
3. Domínguez EC, Boettger D, Cirillo, F, Cabelens F, Eisenach KD, Gagneux S, et al; TBNET; RESIST-TB networks. Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESISTENT-TB consensus statement. Int J Tuberc Lung Dis. 2016 Jan;20(1):24-42. DOI: 10.5588/ijtld.15.0221
4. Brasil. Ministério da Saúde. Departamento de HIV/Aids, Tuberculose, Hepatites Virais e Infecções Sexualmente Transmissíveis. Recomendações técnicas para laudo e interpretação do teste de hibridização em linha (Line Probe Assay - LPA) para tuberculose. Brasília: Ministério da Saúde; 2023 [acesso 2025 mar 20]. Disponível em: https://www.gov.br/aids/pt-br/central-de-conteudo/publicacoes/2023/recomendacoes-tecnicas-para-laudo-e-interpretacao-do-teste-de-hibridizacao-com-sonda-em-linha-line-probe-assay-lpa-para-tuberc.pdf/view
5. Chan ACK, Chan MCH, Yip PCW, Yam WC, Chau CH, Lam RFM, et al. Grave impact of undetected rpoB I572F mutation on clinical course of multidrug-resistant tuberculosis: a case report. Hong Kong Med J. 2023 Feb;29(1):70-2. DOI: 10.12809/hkmj219735
6. Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde. Departamento de Vigilância das Doenças Transmissíveis. Manual de recomendações para o controle da Tuberculose no Brasil. 2ª ed. Brasília: Ministério da Saúde; 2019 [acesso 2025 mar 29]. Disponível em: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/svsa/tuberculose/manual-de-recomendacoes-e-controle-da-tuberculose-no-brasil-2a-ed.pdf/view
7. André E, Goeminne L, Colmant U, Berckert P, Niemann S, Delmee M. Novel rapid PCR or the detection of lle491Phe rpoB mutation of Mycobacterium tuberculosis, a rifampicin-resistance-conferring mutation undetected by commercial assays. Clin Microbiol Infect. 2017 Apr;23(4):267.e5-267.e7. DOI: 10.1016/j.cmi.2016.12.009
8. Alves KKAF, Borralho LM, Araújo AJ, Bernardino IM, Figueiredo TMRM. Fatores associados à cura e ao abandono do tratamento da tuberculose na população privada de liberdade. Rev Bras Epidemiol. 2020;23:e200079. DOI: 10.1590/1980-549720200079
9. Lôbo NMN, Portela MC, Sanchez AAMM. Análise do cuidado em saúde no sistema prisional do Pará, Brasil. Ciênc Saúde Coletiva. 2022 Dez;27(12):4423-34. DOI: 10.1590/1413-812320222712.10212022
10. Rocha MS, Bartholomay P, Cavalcante MV, Medeiros FC, Codenotti SB, Pelissari DM, et al. Notifiable Diseases Information System (SINAN): main features of tuberculosis-related notification and data analysis. Epidemiol Serv Saude. 2020 Feb 17;29(1):e2019017. DOI: 10.5123/S1679-49742020000100009
11. Liu YE, Mabene Y, Camelo S, Rueda ZV, Pelissari DM, Dockhorn Costa Johansen F, et al. Mass incarceration as a driver of the tuberculosis epidemic in Latin America and projected effects of policy alternatives: a mathematical modelling study. Lancet Public Health. 2024 Nov;9(11):e841-e51. DOI: 10.1016/S2468-2667(24)00192-0
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Infections in Evidence

This work is licensed under a Creative Commons Attribution 4.0 International License.
The Copyright Transfer Agreement also understands that the authors guarantee that the respective Case Report has never been published in another communication vehicle or scientific journal. Papers presented at meetings and/or scientific congresses may be published in the electronic Journal INFECTIONS IN EVIDENCE - Case Reports, provided they have not been published in whole or in part in their Proceedings or Annals in the format of a complete article including images, discussion and bibliographic references, or that they have not been assigned a specific DOI number.
















